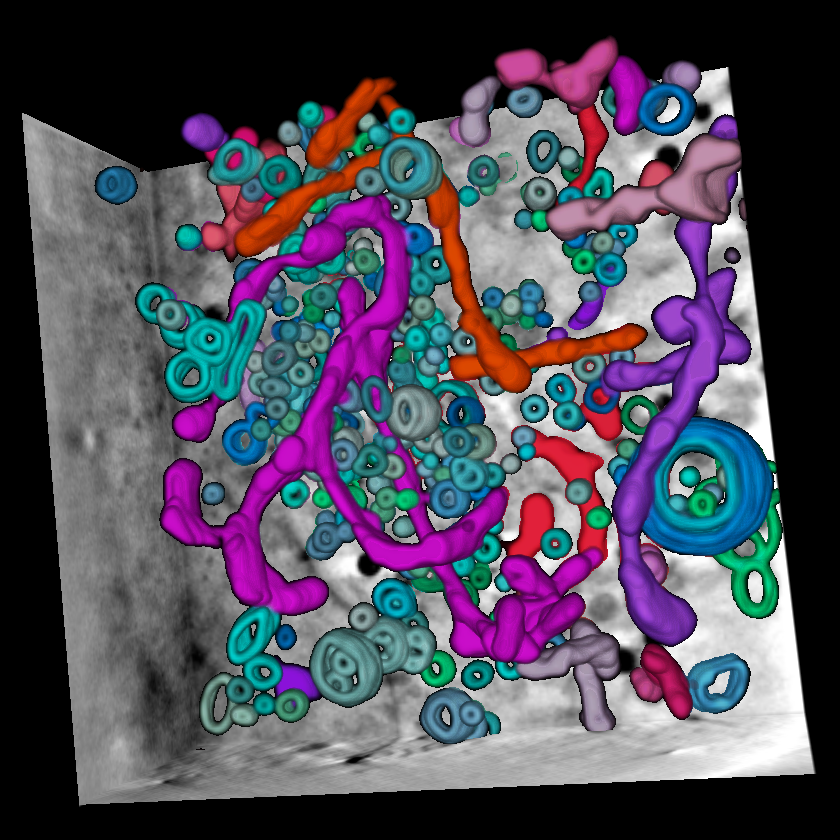
 How does the herpes virus hijack our cells? Cryo-SXT can show us...
How does the herpes virus hijack our cells? Cryo-SXT can show us...Diamond Light Source / PLoS Pathogens
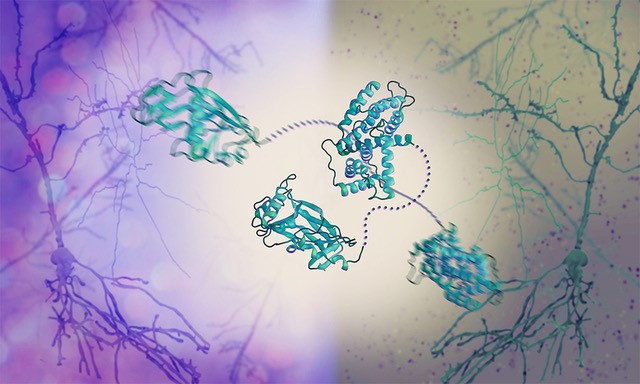 Flexibility is key for herpesvirus hijacking of cellular machinery
Flexibility is key for herpesvirus hijacking of cellular machineryEMBL / PLoS Pathogens
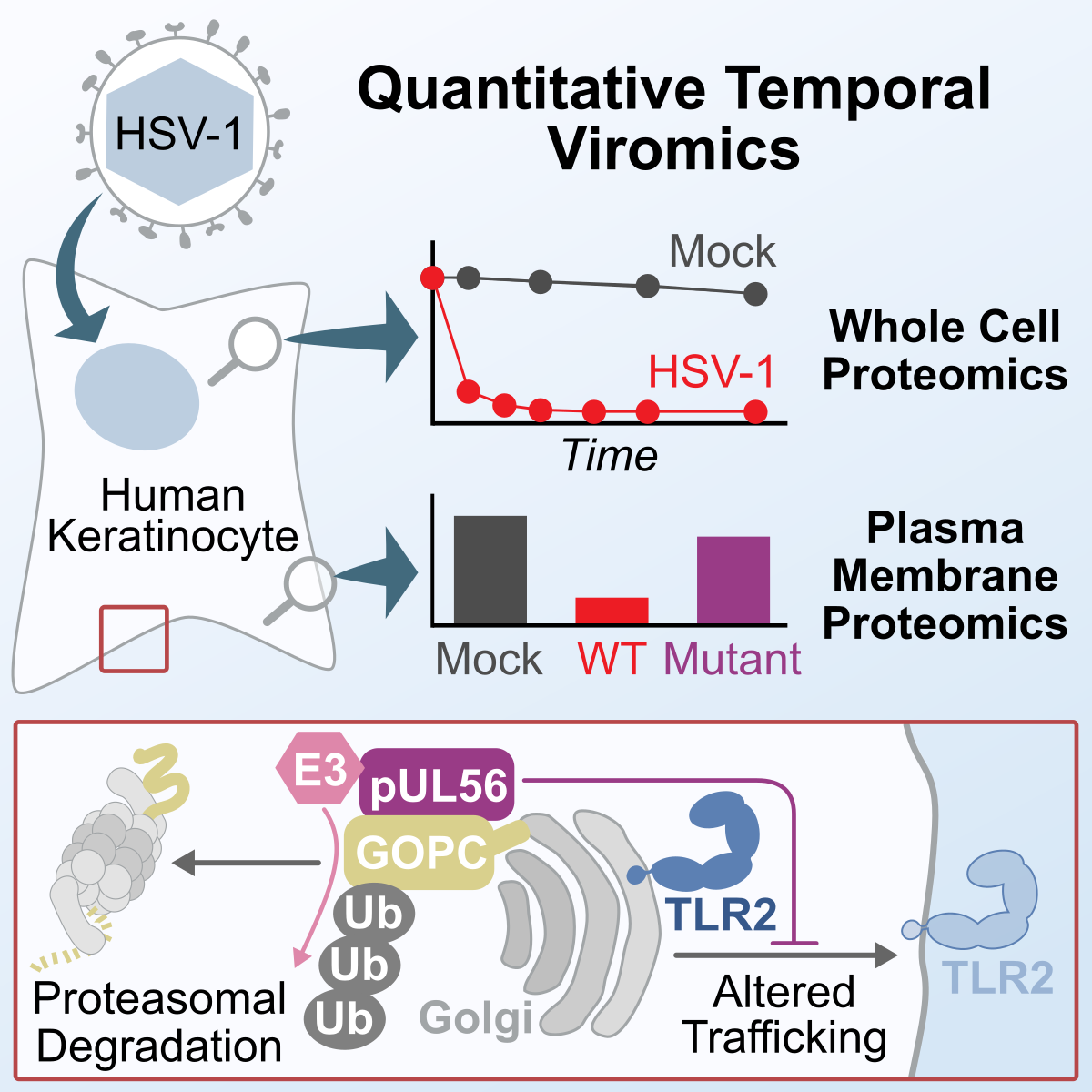 Proteomics of HSV infection
Proteomics of HSV infectionCell Reports
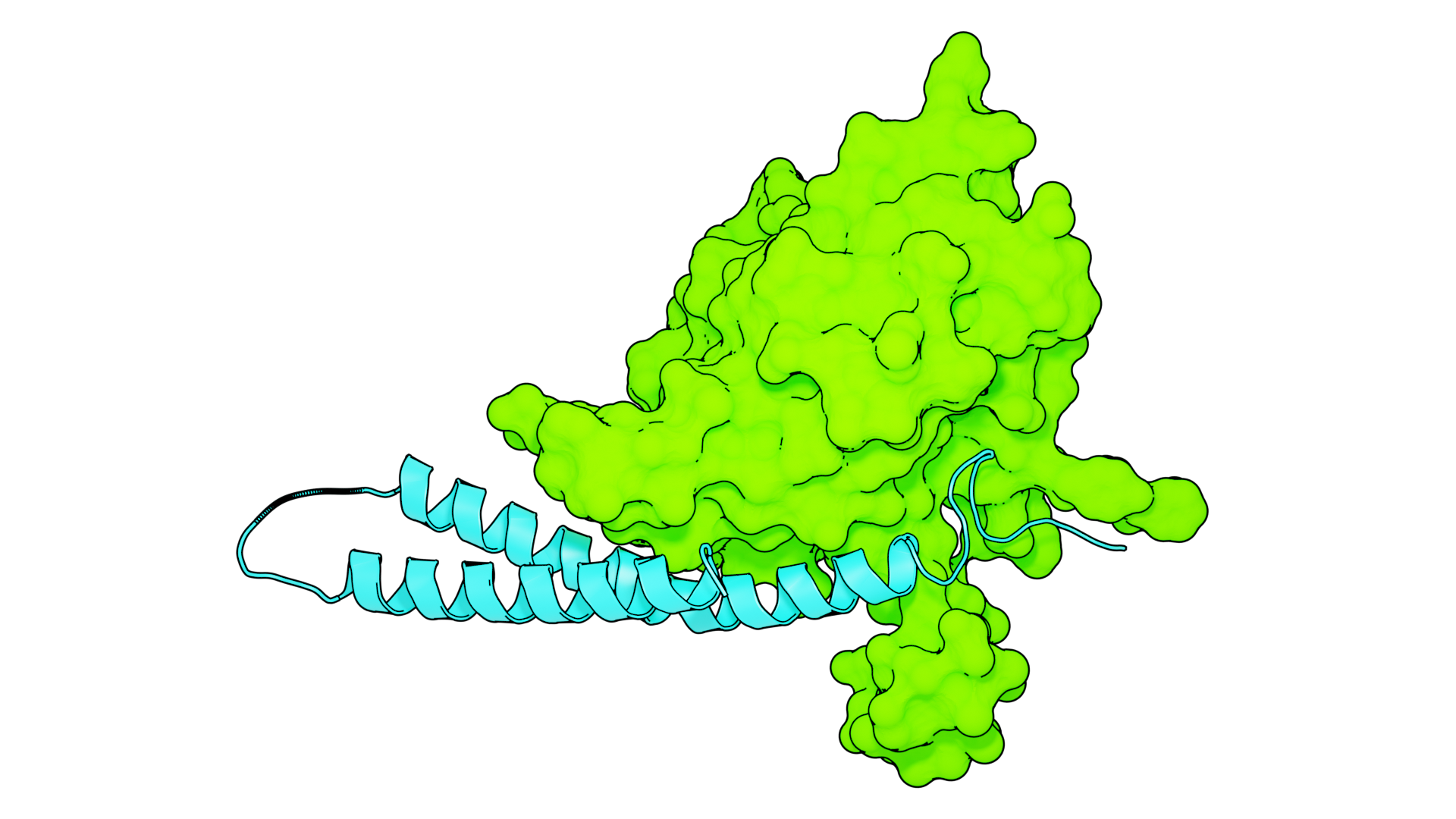 How herpes viruses put on their protective coat
How herpes viruses put on their protective coateLife digest
 The mechanism of sphingolipid processing revealed by a GALC-SapA complex structure
The mechanism of sphingolipid processing revealed by a GALC-SapA complex structureNature Communications
 Cellular and viral peptides bind multiple sites on the N-terminal domain of clathrin
Cellular and viral peptides bind multiple sites on the N-terminal domain of clathrinTraffic
 into the evolution of a complex virus from the crystal structure of vaccinia virus D13
into the evolution of a complex virus from the crystal structure of vaccinia virus D13Structure
Online Preprints
*Joint first authors, ^Joint senior authors- K.L. Nahas, V. Connor, K.J. Wijesinghe, H.G. Barrow, I.M. Dobbie, M. Harkiolaki^, S.C. Graham^, C.M. Crump^ (2024) Applying 3D correlative structured illumination microscopy and X-ray tomography to characterise herpes simplex virus-1 morphogenesis. BioRχiv online preprint doi: 10.1101/2024.03.13.584906
- B.G. Butt*, D. Fischer*, A.R. Rep*, M. Schauflinger, C. Read, T. Böck, M. Hirner, S.C. Graham^, J. von Einem^ (2024) Human cytomegalovirus deploys molecular mimicry to recruit VPS4A to sites of virus assembly. BioRχiv online preprint doi: 10.1101/2024.01.04.572781
- C. Lefévre, G.M. Cook, A.M. Dinan, S. Torii, H. Stewart, G. Gibbons, A.S. Nicholson, L. Echavarria-Consuegra, L.W. Meredith, V. Lulla, J.C. Kenyon, I.G. Goodfellow, J.E. Deane, S.C. Graham, A. Lakatos, L. Lambrechts, I. Brierley, N. Irigoyen (2023) Zika viruses encode multiple upstream open reading frames in the 5' viral region with a role in neurotropism. BioRχiv online preprint doi: 10.1101/112904
Publications
*Joint first authors, ^Joint senior authors- T. Ghosh^, R.G. Almeida, C. Zhao, A. Mannioui, E. Martin, A. Fleet, C.Z. Chen, P. Assinck, S. Ellams, G.A. Gonzalez, S.C. Graham, D.H. Rowitch, K. Stott, I. Adams, B. Zalc, N. Goldman, D.A. Lyons, R.J.M. Franklin^ (2024) A retroviral link to vertebrate myelination through retrotransposon-RNA-mediated control of myelin gene expression. Cell, 187: 814–830.e23 doi: 10.1016/j.cell.2024.01.011
- J. Schmitt, E. Poole, I. Groves, D.J. Owen^, S.C. Graham^, J. Sinclair^, B.T. Kelly^ (2024) Repurposing an endogenous degradation domain for antibody-mediated disposal of cell-surface proteins. EMBO Reports, 25: 951–970 doi: 10.1038/s44319-024-00063-3
- H. Ali, A. Lulla, A.S. Nicholson, J. Hankinson, E.B. Wignall-Fleming, R.L. O'Connor, D.L. Vu, S.C. Graham, J.E. Deane, S. Guix, V. Lulla (2023) Attenuation hotspots in neurotropic human astroviruses. PLoS Biology, 21: e3001815 doi: 10.1371/journal.pbio.3001815
- S.J. McKie, A.S. Nicholson, E. Smith, S. Fawke, E. Caroe, J.C. Williamson, B.G. Butt, D. Kolarov, O. Peterka, M. Holcapek, P.J. Lehner, S.C. Graham, J.E. Deane (2023) Altered plasma membrane abundance of the sulfatide-binding protein NF155 links glycosphingolipid imbalances to demyelination. Proceedings of the National Academy of Sciences of the USA, 120: e2218823120 doi: 10.1073/pnas.2218823120
- N.S. Barbosa, J.O. Concha, L.L.P. da Silva, C.M. Crump^, S.C. Graham^ (2023) Oropouche virus glycoprotein topology and cellular requirements for glycoprotein secretion. Journal of Virology, 97: e01331-22 doi: 10.1128/jvi.01331-22
- I.M. Hay, M. Shamin, E.R. Caroe, A.S.A. Mohammed, D.I. Svergun, C.M Jeffries, S.C. Graham, H.J. Sharpe^, J.E. Deane^ (2023) Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains. Journal of Biological Chemistry, 299: 102750 doi: 10.1016/j.jbc.2022.102750
- T.H. Benedyk, V. Connor, E.R. Caroe, M. Shamin, D.I. Svergun, J.E. Deane, C.M. Jeffries, C.M. Crump, S.C. Graham (2022) Herpes simplex virus 1 protein pUL21 alters ceramide metabolism by activating the inter-organelle transport protein CERT. Journal of Biological Chemistry, 298: 102589 doi: 10.1016/j.jbc.2022.102589
- I.M. Hay, K.E. Mulholland, T. Lai, S.C. Graham, H.J. Sharpe^, J.E. Deane^ (2022) Molecular mechanism of Afadin substrate recruitment to the receptor phosphatase PTPRK via its pseudophosphatase domain. eLife, 11: e79855 doi: 10.7554/eLife.79855
- K.L. Nahas, V. Connor, K.M. Scherer, C.F. Kaminski, M. Harkiolaki^, C.M. Crump^, S.C. Graham^ (2022) Near-native state imaging by cryo-soft-X-ray tomography reveals remodelling of multiple cellular organelles during HSV-1 infection. PLoS Pathogens, 18: e1010629 doi: 10.1371/journal.ppat.1010629
- K.L. Nahas^, J.F. Fernandes, N. Vyas, C.M. Crump, S.C. Graham, M. Harkiolaki^ (2022) Contour: a semi-automated segmentation and quantitation tool for cryo-soft-X-ray tomography. Biological Imaging, 2: e3 doi: 10.1101/2021.12.03.470962
- W.N.D. Gao, C. Gao, J.E. Deane, D.C.J. Carpentier, G.L. Smith, S.C. Graham (2022) The crystal structure of vaccinia virus protein E2 and perspectives on the prediction of novel viral protein folds. Journal of General Virology, 103: 001716 doi: 10.1099/jgv.0.001716
- C.H Hill*^, L. Pekarek*, S. Napthine*, A. Kibe, A.E. Firth, S.C. Graham^, N. Caliskan^, I. Brierley^ (2021) Structural and molecular basis for Cardiovirus 2A protein as a viral gene expression switch. Nature Communications, 12: 7166 doi: 10.1038/s41467-021-27400-7
- M. Shamin, S.J. Spratley, S.C. Graham, J.E. Deane (2021) A tetrameric assembly of saposin A: increasing structural diversity in lipid transfer proteins. Contact, 4: 1–11 doi: 10.1177/25152564211052382
- C.H Hill*^, G.M. Cook*, S. Napthine*, A. Kibe, K. Brown, N. Caliskan, A.E. Firth^, S.C. Graham^, I. Brierley^ (2021) Investigating molecular mechanisms of 2A-stimulated ribosomal pausing and frameshifting in Theilovirus. Nucleic Acids Research, 49: 11938–11958 doi: 10.1093/nar/gkab969
- T.H. Benedyk, J. Muenzner, V. Connor, Y. Han, K. Brown, K.J. Wijesinghe, Y. Zhuang, S. Colaco, G.A. Stoll, O.S. Tutt, S. Svobodova, D.I. Svergun, N.A. Bryant, J.E. Deane, A.E. Firth, C.M. Jeffries, C.M. Crump^, S.C. Graham^ (2021) pUL21 is a viral phosphatase adaptor that promotes herpes simplex virus replication and spread. PLoS Pathogens, 17: e1009824 doi: 10.1371/journal.ppat.1009824
- G. Gallo, C. Conceicao, C. Tsirigoti, B. Willett, S.C. Graham, D. Bailey (2021) Application of error-prone PCR to functionally probe the morbillivirus Haemagglutinin protein. Journal of General Virology, 102: 001580 doi: 10.1099/jgv.0.001580
- C. Conceicao*, N. Thakur*, S. Human, J.T. Kelly, L. Logan, D. Bialy, S. Bhat, P. Stevenson-Leggett, A.K. Zagrajek, P. Hollinghurst, M. Varga, C. Tsirigoti, M. Tully, C. Chiu, K. Moffat, A.P. Silesian, J.A. Hammond, H.J. Maier, E. Bickerton, H. Shelton, I. Dietrich, S.C. Graham, D. Bailey (2020) The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biology, 18: e3001016 doi: 10.1371/journal.pbio.3001016
- T.K. Soh*, C.T.R. Davies*, J. Muenzner*, L.M. Hunter, H.G. Barrow, V. Connor, C.R. Bouton, C. Smith, E. Emmott, R. Antrobus, S.C. Graham^, M.P. Weekes^, C.M. Crump^ (2020) Temporal Proteomic Analysis of Herpes Simplex Virus 1 Infection Reveals Cell-Surface Remodeling via pUL56-Mediated GOPC Degradation. Cell Reports, 33: 108235 doi: 10.1016/j.celrep.2020.108235
- B.G. Butt, E.J. Scourfield, S.C. Graham (2020) Non-native fold of the putative VPS39 zinc finger domain. [version 2; peer review: 2 approved]. Wellcome Open Research, 5: 154 doi: 10.12688/wellcomeopenres.16078.2
- B.G. Butt, D.J. Owen, C.M. Jeffries, L. Ivanova, C.H. Hill, J.W. Houghton, M.F. Ahmed, R. Antrobus, D.I. Svergun, J.J. Welch, C.M. Crump, S.C. Graham (2020) Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex. eLife, 9: e53789 doi: 10.7554/eLife.53789
- M. Shamin, T.H. Benedyk, S.C. Graham, J.E. Deane (2019) The lipid transfer protein Saposin B does not directly bind CD1d for lipid antigen loading. [version 2; peer review: 3 approved] Wellcome Open Research, 4: 117 doi: 10.12688/wellcomeopenres.15368.2
- S.C. Graham, B. Nagar, G.G. Privé, J.E. Deane (2019) Molecular models should not be published without the corresponding atomic coordinates. Proceedings of the National Academy of Sciences of the USA, 116: 11099–11100 doi: 10.1073/pnas.1904409116
- C. Gao, M.A. Pallett, T.I. Croll, G.L. Smith, S.C. Graham (2019) Molecular basis of cullin-3 (Cul3) ubiquitin ligase subversion by vaccinia virus protein A55. Journal of Biological Chemistry, 294: 6416–6429 doi: 10.1074/jbc.RA118.006561
- J.M. Alves, M. Carneiro, J.Y. Cheng, A. Lemos de Matos, M.M. Rahman, L. Loog, P.F. Campos, N. Wales, A. Eriksson, A. Manica, T. Strive, S.C. Graham, S. Afonso, D.J. Bell, L. Belmont, J.P. Day, S.J. Fuller, S. Marchandeau, W.J. Palmer, G. Queney, A.K. Surridge, F.G. Vieira, G. McFadden, R. Nielsen, M.T.P. Gilbert, P.J. Esteves, N. Ferrand, F.M. Jiggins (2019) Parallel adaptation of rabbit populations to myxoma virus. Science, 363: 1319–1326 doi: 10.1126/science.aau7285
- N. Abdullah, J.T. Kelly, S.C. Graham, J. Birch, D. Gonçalves-Carneiro, T. Mitchell, R. N. Thompson, K.A. Lythgoe, N. Logan, M.J. Hosie, V.N. Bavro, B.J. Willett, M.P. Heaton, D. Bailey (2018) Structure-guided identification of a non-human morbillivirus with zoonotic potential. Journal of Virology, 23: e01248-18 doi: 10.1128/JVI.01248-18
- M.R. Hunter, G.G. Hesketh, T.H. Benedyk, A.C. Gingras, S.C. Graham (2018) Proteomic and biochemical comparison of the cellular interaction partners of human VPS33A and VPS33B. Journal of Molecular Biology, 430: 2153–2163 doi: 10.1016/j.jmb.2018.05.019
- R.C. Fleith*, H.V. Mears*, X.Y. Leong, T.J. Sanford, E. Emmott, S.C. Graham, D.S. Mansur, T.R. Sweeney (2018) IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA. Nucleic Acids Research, 46: 5269–5285 doi: 10.1093/nar/gky191
- C.H. Hill, G.M. Cook, S.J. Spratley, S.C. Graham, J.E. Deane (2018) The mechanism of sphingolipid processing revealed by a GALC-SapA complex structure. Nature Communications, 9: 151 doi: 10.1038/s41467-017-02361-y
- M.R. Hunter*, E.J. Scourfield*, E. Emmott, S.C. Graham (2017) VPS18 recruits VPS41 to the human HOPS complex via a RING-RING interaction. Biochemical Journal, 474: 3615–3626 doi: 10.1042/BCJ20170588
- A. Albecka, D.J. Owen, L. Ivanova, J. Brun, R. Liman, L. Davies, M.F. Ahmed, S. Colaco, M. Hollinshead, S.C. Graham^ and C.M. Crump^ (2017) Dual function of the pUL7-pUL51 tegument protein complex in HSV-1 infection. Journal of Virology, 91: e02196-16 doi: 10.1128/JVI.02196-16
- J. Muenzner, L.M. Traub, B.T. Kelly^, S.C. Graham^ (2017) Cellular and viral peptides bind multiple sites on the N-terminal domain of clathrin. Traffic, 18: 44–57 doi: 10.1111/tra.12457
- B.T. Kelly, S.C. Graham, D.J. Owen (2016) Using selenomethionyl derivatives to assign sequence in low-resolution structures of the AP2 clathrin adaptor. Acta Crystallographica Section D: Biological Crystallography, 72: 336–345 doi: 10.1107/S2059798315021580
- E.N. Leen, F. Sorgeloos, S. Correia, Y. Chaudhry, F. Cannac, C. Pastore, Y. Xu, S.C. Graham, S.J. Matthews, I.G. Goodfellow, S. Curry (2016) A conserved interaction between a C-terminal motif in Norovirus VPg and the HEAT-1 domain of eIF4G is essential for translation initiation. PLoS Pathogens, 12: e1005379 doi: 10.1371/journal.ppat.1005379
- D.J. Owen, C.M. Crump, S.C. Graham (2015) Tegument Assembly and Secondary Envelopment of Alphaherpesviruses. Viruses, 7: 5084–5114 doi: 10.3390/v7092861
- L. Wartosch, U. Günesdogan, S.C. Graham, J.P. Luzio (2015) Recruitment of VPS33A to HOPS by VPS16 is required for lysosome fusion with endosomes and autophagosomes. Traffic, 16: 727–742 doi: 10.1111/tra.12283
- S. Neidel, C. Maluquer de Motes, D.S. Mansur, P. Strnadova, G.L. Smith, S.C. Graham (2015) Vaccinia Virus Protein A49 is an Unexpected Member of the B-cell Lymphoma (Bcl)-2 Protein Family. Journal of Biological Chemistry, 290: 5991–6002 doi: 10.1074/jbc.M114.624650
- B.T. Kelly, S.C. Graham, N. Liska, P.N. Dannhauser, S. Höning, E.J. Ungewickell, D.J. Owen (2014) AP2 controls clathrin polymerization with a membrane-activated switch. Science, 345: 459–463 doi: 10.1126/science.1254836
- C.H. Hill, S.C. Graham, R.J. Read, J.E. Deane (2013) Structural snapshots illustrate the catalytic cycle of β-galactocerebrosidase, the defective enzyme in Krabbe disease. Proceedings of the National Academy of Sciences of the USA, 110: 20479–20484 doi: 10.1073/pnas.1311990110
- Y. Hackmann, S.C. Graham^, S. Ehl, S. Höning, K. Lehmberg, M. Aricò, D.J. Owen, G.M. Griffiths^ (2013) Syntaxin binding mechanism and disease-causing mutations in Munc18-2. Proceedings of the National Academy of Sciences of the USA, 110: E4482–E4491 doi: 10.1073/pnas.1313474110
- S.C. Graham*^, L. Wartosch*, S.R. Gray, E.J. Scourfield, J.E. Deane, J.P. Luzio^, D.J. Owen (2013) Structural basis of Vps33A recruitment to the human HOPS complex by Vps16. Proceedings of the National Academy of Sciences of the USA, 110: 13345–13350 doi: 10.1073/pnas.1307074110
- M.W. Bahar, L.P. Sarin, S.C. Graham, J. Pang, D.H. Bamford, D.I. Stuart, J.M. Grimes (2013) Structure of a VP1-VP3 complex suggests how birnaviruses package the VP1 polymerase. Journal of Virology, 87: 3229–3236 doi: 10.1128/JVI.02939-12
- C. Maluquer de Motes, S. Cooray, K. McGourty, H. Ren, M.W. Bahar, D.I. Stuart, J.M. Grimes, S.C. Graham, G.L. Smith (2011) Inhibition of apoptosis and NF-κB activation by vaccinia protein N1 occur via distinct binding surfaces and make different contributions to virulence. PLoS Pathogens, 7: e1002430 doi: 10.1371/journal.ppat.1002430
- S.E. Miller*, D. Sahlender*, S.C. Graham, S Höning, M.S. Robinson, A.A. Peden, D.J. Owen (2011) The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell, 147: 1118–1131 doi: 10.1016/j.cell.2011.10.038
- J.E. Deane, S.C. Graham, N.N. Kim, P.E. Stein, R. McNair, M.B. Cachón-González, T.M. Cox, R.J. Read (2011) Insights into Krabbe Disease from the Structure of Galactocerebrosidase. Proceedings of the National Academy of Sciences of the USA, 108: 15169–15173 doi: 10.1073/pnas.1105639108
- M.W. Bahar, S.C. Graham, D.I. Stuart, J.M. Grimes (2011) Insights into the evolution of a complex virus from the crystal structure of vaccinia virus D13. Structure, 19: 1011–1020 doi: 10.1016/j.str.2011.03.023
- M.W. Bahar*, S.C. Graham*, R. Chen, S. Cooray, G.L. Smith, D.I. Stuart, J.M. Grimes (2011) How vaccinia virus has evolved to subvert the host immune response. Journal of Structural Biology, 175: 127–134 doi: 10.1016/j.jsb.2011.03.010
- S.C. Graham*, L.P. Sarin*, M.W. Bahar, R.A. Myers, D.I. Stuart, D.H. Bamford, J.M. Grimes (2011) The N-terminus of the RNA polymerase from Infectious Pancreatic Necrosis Virus is the determinant of genome attachment. PLoS Pathogens, 7: e1002085 doi: 10.1371/journal.ppat.1002085
- C.T. Benfield, D. Mansur, L.E. McCoy, B.J. Ferguson, M.W. Bahar, A.P. Oldring, J.M. Grimes, D. I. Stuart, S.C. Graham^, G.L. Smith^ (2011) Mapping the IκB kinase beta (IKKβ)-binding interface of B14, a vaccinia virus inhibitor of IKKβ-mediated activation of nuclear factor κB. Journal of Biological Chemistry, 286: 20727–20735 doi: 10.1074/jbc.M111.231381
- D. Bubeck, M.A.M. Reijns, S.C. Graham, K.R. Astell, E.Y. Jones, A.P. Jackson (2011) PCNA directs Type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Research, 39: 3652–3366 doi: 10.1093/nar/gkq980
- M.A.M. Reijns*, D. Bubeck*, L.C. Gibson, S.C. Graham, G.S. Baillie, E.Y. Jones, A.P. Jackson (2011) The structure of the human RNase H2 complex defines key interaction interfaces relevant to enzyme function and human disease. Journal of Biological Chemistry, 286: 10530–10539 doi: 10.1074/jbc.M110.177394
- R. Assenberg, O. Delmas, B. Morin, S.C. Graham, X. De Lamballerie, C. Laubert, B. Coutard, J.M. Grimes, J. Neyts, R.J. Owens, B.W. Brandt, A. Gorbalenya, P. Tucker, D.I. Stuart, B. Canard, H. Bourhy (2010) Genomics and structure/function studies of Rhabdoviridae proteins involved in replication and transcription. Antiviral Research, 87: 149–161 doi: 10.1016/j.antiviral.2010.02.322
- R. Assenberg, O. Delmas, J. Ren, P.O. Vidalain, A. Verma, F. Larrous, S.C. Graham, F. Tangy, J.M. Grimes, H. Bourhy (2010) The Structure of the N-RNA Binding Domain of the Mokola virus Phosphoprotein. Journal of Virology, 84: 1089–1096 doi: 10.1128/JVI.01520-09
- D. Hatherley, S.C. Graham, K. Harlos, D.I. Stuart, A.N. Barclay (2009) Structure of signal regulatory protein α: a link to antigen receptor evolution. Journal of Biological Chemistry, 284: 26613–26619 doi: 10.1074/jbc.M109.017566
- T.A. Bowden, M. Crispin, S.C. Graham, D.J. Harvey, J.M. Grimes, E.Y. Jones, D.I. Stuart (2009) Unusual Molecular Architecture of the Machupo Virus Attachment Glycoprotein. Journal of Virology, 83: 8259–8265 doi: 10.1128/JVI.00761-09
- B.L. Walter, A.E. Armitage, S.C. Graham, T. de Oliveira, P. Skinhøj, E.Y. Jones, D.I. Stuart, A.J. McMichael, B. Chesebro, A.K. Iversen (2009) Functional characteristics of HIV-1 subtype C compatible with increased heterosexual transmissibility. AIDS, 23: 1047–1057 doi: 10.1097/QAD.0b013e32832a1806
- S.C. Graham, R. Assenberg*, O. Delmas*, A. Verma, A. Gholami, C. Talbi, R.J. Owens, D.I. Stuart, J.M. Grimes, H. Bourhy (2008) Rhabdovirus matrix protein structures reveal a novel mode of self-association. PLoS Pathogens, 4: e1000251 doi: 10.1371/journal.ppat.1000251
- S.C. Graham*, M.W. Bahar*, S. Cooray*, R.A.J. Chen, D.M. Whalen, N.G.A. Abrescia, D. Alderton, R.J. Owens, D.I. Stuart, G.L. Smith, J.M. Grimes (2008) Vaccinia virus proteins A52 and B14 share a Bcl-2−like fold but have evolved to inhibit NF-κB rather than apoptosis. PLoS Pathogens, 4: e1000128 doi: 10.1371/journal.ppat.1000128
- D. Hatherley*, S.C. Graham*, K. Harlos, J. Turner, D.I. Stuart, A.N. Barclay (2008) Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47. Molecular Cell, 31: 266–277 doi: 10.1016/j.molcel.2008.05.026
- J.E. Deane, S.C. Graham, E.P. Mitchell, D. Flot, S. Johnson, S.M. Lea (2008) Crystal structure of Spa40, the specificity switch for the Shigella flexneri Type III Secretion System. Molecular Microbiology, 69: 267–276 doi: 10.1111/j.1365-2958.2008.06293.x
- R. Assenberg, O. Delmas, S.C. Graham, A. Verma, N. Berrow, D.I. Stuart, R.J. Owens, H. Bourhy, J.M. Grimes (2008) Expression, purification and crystallization of a lyssavirus matrix (M) protein. Acta Crystallographica Section F: Structural Biology and Crystallization Communications, 64: 258–262 doi: 10.1107/S1744309108004557
- S.C. Graham, J.M. Guss (2008) Complexes of mutants of Escherichia coli aminopeptidase P and the tripeptide substrate ValProLeu. Archives of Biochemistry and Biophysics, 469: 200–208 doi: 10.1016/j.abb.2007.10.009
- T.S. Walter, E.J. Mancini, J. Kadlec, S.C. Graham, R. Assenberg, J. Ren, S. Sainsbury, R.J. Owens, D.I. Stuart, J.M. Grimes, K. Harlos (2008) Semi-Automated Microseeding of Nanolitre Crystallization Experiments. Acta Crystallographica Section F: Structural Biology and Crystallization Communications, 64: 14–18 doi: 10.1107/S1744309107057260
- S.C. Graham, M.W. Bahar, N.G.A. Abrescia, G.L. Smith, D.I. Stuart, J.M. Grimes (2007) Structure of CrmE, a Virus-encoded Tumour Necrosis Factor Receptor. Journal of Molecular Biology, 372: 660–671 doi: 10.1016/j.jmb.2007.06.082
- M. Lee, C.W. Chan, S.C. Graham, R.I. Christopherson, J.M. Guss, M.J. Maher (2007) Structures of Ligand-free and Inhibitor Complexes of Dihydroorotase from Escherichia coli: Implications for Loop Movement in Inhibitor Design. Journal of Molecular Biology, 370: 812–825 doi: 10.1016/j.jmb.2007.05.019
- C.M. Jeffries, S.C. Graham, P.H. Stokes, C.A. Collyer, J.M. Guss, J.M. Matthews (2006) Stabilization of a binary protein complex by intein-mediated circularization. Protein Science, 15: 2612–2618 doi: 10.1110/ps.062377006
- S.C. Graham, P.E. Lilley, M. Lee, P.M. Schaeffer, A.V. Kralicek, N.E. Dixon, J.M. Guss (2006) Kinetic and crystallographic analysis of mutant Escherichia coli aminopeptidase P: Insights into substrate recognition and the mechanism of catalysis. Biochemistry, 45: 964–975 doi: 10.1021/bi0518904
- S.C. Graham, C.S. Bond, H.C. Freeman, J.M. Guss (2005) Structural and functional implications of metal ion selection in aminopeptidase P, a metalloprotease with a dinuclear metal centre. Biochemistry, 44: 13820–13836 doi: 10.1021/bi0512849
- S.C. Graham, M.J. Maher, W.H. Simmons, H.C. Freeman, J.M. Guss (2004) Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin. Acta Crystallographica Section D: Biological Crystallography, 60: 1770–1779 doi: 10.1107/S0907444904018724
- D.B. Langley, D.W.S. Harty, S.C. Graham, J.M. Guss, N. Hunter, C. Collyer (2004) Crystallization of GcnA, an N-acetyl-β-D-glucosaminidase, from Streptococcus gordonii. Acta Crystallographica Section D: Biological Crystallography, 60: 1910–1911 doi: 10.1107/S0907444904019572
- J.E. Deane, M.J. Maher, D.B. Langley, S.C. Graham, J.E. Visvader, J.M. Guss, J.M. Matthews (2003) Crystallization of FLINC4, an intra-molecular LMO4:ldb1 complex. Acta Crystallographica Section D: Biological Crystallography, 59: 1484–1486 doi: 10.1107/S0907444903011843
- S.C. Graham, M. Lee, H.C. Freeman, J.M. Guss (2003) An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 Å resolution. Acta Crystallographica Section D: Biological Crystallography, 59: 897–902 doi: 10.1107/S0907444903005870
- W.M. Shui, R.K. Wong, S.C. Graham, L. Lee, W.B. Church (2003) Integrating, Managing and Analyzing Protein Structures with XML Databases. Proceedings of the 8th International Conference on Database Systems for Advanced Applications, 319–326 doi: 10.1109/DASFAA.2003.1192397
- W.M. Shui, R.K. Wong, S.C. Graham, L. Lee, W.B. Church (2003) A new approach to protein structure and function analysis using semi-structured databases. Proceedings of the First Asia Pacific Bioinformatics Conference, 61–69 http://crpit.com/confpapers/CRPITV19Shui.pdf
- R.K. Wong, F. Lam, S. Graham, W. Shui (2000) An XML Repository for Molecular Sequence Data. Proceedings of the First IEEE International Symposium on Bio-Informatics and Biomedical Engineering, 35–42 doi: 10.1109/BIBE.2000.889587
